Difference between revisions of "AlsD"
| Line 29: | Line 29: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=alsD_3708799_3709566_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:alsD_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=alsD_3708799_3709566_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:alsD_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU36000]] |
|- | |- | ||
|} | |} | ||
Revision as of 14:44, 16 May 2013
- Description: acetolactate decarboxylase
| Gene name | alsD |
| Synonyms | |
| Essential | no |
| Product | acetolactate decarboxylase) |
| Function | overflow metabolism |
| Gene expression levels in SubtiExpress: alsD | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 28 kDa, 4.603 |
| Gene length, protein length | 765 bp, 255 aa |
| Immediate neighbours | ywrO, alsS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 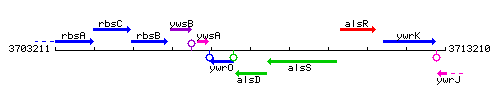
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
AlsR regulon, Rex regulon, stringent response
The gene
Basic information
- Locus tag: BSU36000
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (2S)-2-hydroxy-2-methyl-3-oxobutanoate = (3R)-3-hydroxybutan-2-one + CO2 (according to Swiss-Prot)
- Protein family: alpha-acetolactate decarboxylase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylated on ser/ thr/ tyr PubMed, in vitro phosphorylated by PrkC on Ser-88 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: Q04777
- KEGG entry: [3]
- E.C. number: 4.1.1.5
Additional information
Expression and regulation
- Regulation:
Note: since acetate formation requires ackA activation by CcpA there is an indirect effect of CcpA on the alsSD operon: the operon is not expressed in ccpA mutants
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Nico Pietack, Dörte Becher, Sebastian R Schmidl, Milton H Saier, Michael Hecker, Fabian M Commichau, Jörg Stülke
In vitro phosphorylation of key metabolic enzymes from Bacillus subtilis: PrkC phosphorylates enzymes from different branches of basic metabolism.
J Mol Microbiol Biotechnol: 2010, 18(3);129-40
[PubMed:20389117]
[WorldCat.org]
[DOI]
(I p)
Shigeo Tojo, Kanako Kumamoto, Kazutake Hirooka, Yasutaro Fujita
Heavy involvement of stringent transcription control depending on the adenine or guanine species of the transcription initiation site in glucose and pyruvate metabolism in Bacillus subtilis.
J Bacteriol: 2010, 192(6);1573-85
[PubMed:20081037]
[WorldCat.org]
[DOI]
(I p)
Takashi Inaoka, Takenori Satomura, Yasutaro Fujita, Kozo Ochi
Novel gene regulation mediated by overproduction of secondary metabolite neotrehalosadiamine in Bacillus subtilis.
FEMS Microbiol Lett: 2009, 291(2);151-6
[PubMed:19087206]
[WorldCat.org]
[DOI]
(I p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Heike Reents, Richard Münch, Thorben Dammeyer, Dieter Jahn, Elisabeth Härtig
The Fnr regulon of Bacillus subtilis.
J Bacteriol: 2006, 188(3);1103-12
[PubMed:16428414]
[WorldCat.org]
[DOI]
(P p)
A J Turinsky, T R Moir-Blais, F J Grundy, T M Henkin
Bacillus subtilis ccpA gene mutants specifically defective in activation of acetoin biosynthesis.
J Bacteriol: 2000, 182(19);5611-4
[PubMed:10986270]
[WorldCat.org]
[DOI]
(P p)
M C Renna, N Najimudin, L R Winik, S A Zahler
Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin.
J Bacteriol: 1993, 175(12);3863-75
[PubMed:7685336]
[WorldCat.org]
[DOI]
(P p)