Difference between revisions of "CitZ"
(→Biological materials) |
(→Biological materials) |
||
| Line 148: | Line 148: | ||
** 1A999 ( ''citZ''::''spec''), {{PubMed| }}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A999&Search=1A999 BGSC] | ** 1A999 ( ''citZ''::''spec''), {{PubMed| }}, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A999&Search=1A999 BGSC] | ||
** GP790 (''[[citZ]]-[[icd]]-[[mdh]]''::''kan''), available in [[Jörg Stülke]]'s lab | ** GP790 (''[[citZ]]-[[icd]]-[[mdh]]''::''kan''), available in [[Jörg Stülke]]'s lab | ||
| − | ** GP797 ( ''citZ''::''spec''), allows expression of [['' | + | ** GP797 ( ''citZ''::''spec''), allows expression of ''[[icd]]'' and ''[[mdh]]'', available in [[Jörg Stülke]]'s lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
Revision as of 13:44, 8 December 2014
- Description: citrate synthase
| Gene name | citZ |
| Synonyms | citA2 |
| Essential | no |
| Product | citrate synthase II |
| Function | TCA cycle |
| Gene expression levels in SubtiExpress: citZ | |
| Interactions involving this protein in SubtInteract: CitZ | |
| Metabolic function and regulation of this protein in SubtiPathways: citZ | |
| MW, pI | 41 kDa, 5.451 |
| Gene length, protein length | 1116 bp, 372 aa |
| Immediate neighbours | icd, ytwI |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 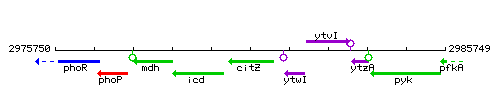
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29140
Phenotypes of a mutant
- glutamate auxotrophy and a defect in sporulation PubMed
Database entries
- BsubCyc: BSU29140
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Acetyl-CoA + H2O + oxaloacetate = citrate + CoA (according to Swiss-Prot)
- Protein family: citrate synthase family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Modification: phosphorylation on Ser-284 PubMed
- Effectors of protein activity:
- Inhibited by acetyl-CoA, 2-oxoglutarate and NADH PubMed FEBS Letters
- Inhibited by citrate and CoA (competitively against acetyl-CoA and non-competitively against oxaloacetate) PubMed
- Inhibited by ATP competitively in B. subtilis strain 168 and HS 1A17 PubMed PubMed
- In B. subtilis strain HS 2A2, ATP inhibits a non-competitive fashion PubMed
- Activated by AMP PubMed
- Localization:
- cytoplasm (homogeneously distributed throughout the cell) PubMed
Database entries
- BsubCyc: BSU29140
- Structure:
- UniProt: P39120
- KEGG entry: [3]
- E.C. number: 2.3.3.1
Additional information
- extensive information on the structure and enzymatic properties of CitZ can be found at Proteopedia
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 8373 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 20578 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 22342 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 10224 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 18693 PubMed
Biological materials
- Mutant:
- GP678 (erm), GP797 (spec) available in Jörg Stülke's lab
- 1A999 ( citZ::spec), PubMed, available at BGSC
- GP790 (citZ-icd-mdh::kan), available in Jörg Stülke's lab
- GP797 ( citZ::spec), allows expression of icd and mdh, available in Jörg Stülke's lab
- Expression vector:
- pGP1120 (N-terminal Strep-tag, for SPINE, purification from B. subtilis, in pGP380) (available in Jörg Stülke's lab)
- pGP1776 (for expression, purification in E. coli with N-terminal Strep-tag, in pGP172, available in Jörg Stülke's lab)
- pGP1761 (expression with N-terminal His-tag from E. coli, in pWH844), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody: available in Linc Sonenshein's lab
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Original publications
Leigh G Monahan, Isabella V Hajduk, Sinead P Blaber, Ian G Charles, Elizabeth J Harry
Coordinating bacterial cell division with nutrient availability: a role for glycolysis.
mBio: 2014, 5(3);e00935-14
[PubMed:24825009]
[WorldCat.org]
[DOI]
(I e)
Michael Kohlstedt, Praveen K Sappa, Hanna Meyer, Sandra Maaß, Adrienne Zaprasis, Tamara Hoffmann, Judith Becker, Leif Steil, Michael Hecker, Jan Maarten van Dijl, Michael Lalk, Ulrike Mäder, Jörg Stülke, Erhard Bremer, Uwe Völker, Christoph Wittmann
Adaptation of Bacillus subtilis carbon core metabolism to simultaneous nutrient limitation and osmotic challenge: a multi-omics perspective.
Environ Microbiol: 2014, 16(6);1898-917
[PubMed:24571712]
[WorldCat.org]
[DOI]
(I p)
Kieran B Pechter, Frederik M Meyer, Alisa W Serio, Jörg Stülke, Abraham L Sonenshein
Two roles for aconitase in the regulation of tricarboxylic acid branch gene expression in Bacillus subtilis.
J Bacteriol: 2013, 195(7);1525-37
[PubMed:23354745]
[WorldCat.org]
[DOI]
(I p)
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Agnes Roux, Abraham L Sonenshein
Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes.
Mol Microbiol: 2002, 45(1);179-90
[PubMed:12100558]
[WorldCat.org]
[DOI]
(P p)
C Jourlin-Castelli, N Mani, M M Nakano, A L Sonenshein
CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis.
J Mol Biol: 2000, 295(4);865-78
[PubMed:10656796]
[WorldCat.org]
[DOI]
(P p)
K Matsuno, T Blais, A W Serio, T Conway, T M Henkin, A L Sonenshein
Metabolic imbalance and sporulation in an isocitrate dehydrogenase mutant of Bacillus subtilis.
J Bacteriol: 1999, 181(11);3382-91
[PubMed:10348849]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, P Zuber, A L Sonenshein
Anaerobic regulation of Bacillus subtilis Krebs cycle genes.
J Bacteriol: 1998, 180(13);3304-11
[PubMed:9642180]
[WorldCat.org]
[DOI]
(P p)
S Jin, A L Sonenshein
Characterization of the major citrate synthase of Bacillus subtilis.
J Bacteriol: 1996, 178(12);3658-60
[PubMed:8655569]
[WorldCat.org]
[DOI]
(P p)
S Jin, A L Sonenshein
Transcriptional regulation of Bacillus subtilis citrate synthase genes.
J Bacteriol: 1994, 176(15);4680-90
[PubMed:8045899]
[WorldCat.org]
[DOI]
(P p)
S Jin, A L Sonenshein
Identification of two distinct Bacillus subtilis citrate synthase genes.
J Bacteriol: 1994, 176(15);4669-79
[PubMed:8045898]
[WorldCat.org]
[DOI]
(P p)
D E Johnson, R S Hanson
Bacterial citrate synthases: purification, molecular weight and kinetic mechanism.
Biochim Biophys Acta: 1974, 350(2);336-53
[PubMed:4211224]
[WorldCat.org]
[DOI]
(P p)
V R Flechtner, R S Hanson
Coarse and fine control of citrate synthase from Bacillus subtilis.
Biochim Biophys Acta: 1969, 184(2);252-62
[PubMed:4980242]
[WorldCat.org]
[DOI]
(P p)