Difference between revisions of "PtsI"
| Line 22: | Line 22: | ||
|style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ptsI_nucleotide.txt Gene sequence (+200bp) ]''' | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ptsI_nucleotide.txt Gene sequence (+200bp) ]''' | ||
|style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ptsI_protein.txt Protein sequence]''' | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/ptsI_protein.txt Protein sequence]''' | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;color:#FF0000" align="center" | '''Caution: The sequence for this gene in SubtiList contains errors | ||
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ptsI_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ptsI_context.gif]] | ||
Revision as of 14:11, 23 February 2009
- Description: Enzyme I, general (non sugar-specific) component of the PTS. Enzyme I transfers the phosphoryl group from phosphoenolpyruvate (PEP) to the phosphoryl carrier protein (HPr)
| Gene name | ptsI |
| Synonyms | |
| Essential | no |
| Product | phosphotransferase system (PTS) enzyme I |
| Function | PTS-dependent sugar transport |
| MW, pI | 62,9 kDa, 4.59 |
| Gene length, protein length | 1710 bp, 570 amino acids |
| Immediate neighbours | ptsH, splA |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 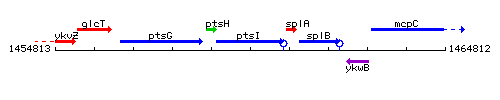
| |
Contents
The gene
Basic information
- Coordinates: 1458959 - 1460668
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: PEP-dependent autophosphorylation on His-189, transfer of the phosphoryl group to HPr (His-15)
- Protein family: PEP-utilizing enzyme family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- HPr binding site (N-Terminal Domain)
- pyruvate binding site (C-Terminal Domain)
- pyrophosphate/phosphate carrier histidine (central Domain)
- Modification: transient autophosphorylation on His-189, in vivo also phosphorylated on Ser-44 or Ser-46 PubMed
- Cofactor(s): Magnesium
- Effectors of protein activity:
- Interactions:
- Localization: Cytoplasm
Database entries
- Structure:
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant: GP864 (ermC), available in Stülke lab
- Expression vector: pAG3 (His-tag), available in Galinier lab
- lacZ fusion:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Josef Deutscher, Paris-Grignon, France
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
- Frisby, D., and Zuber, P. 1994. Mutations in pts cause catabolite-resistant sporulation and altered regulation of spo0H in Bacillus subtilis. J. Bacteriol. 176: 2587-2595. PubMed
- Macek B, Mijakovic I, Olsen JV (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics 6(4): 697-707. PubMed