Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' cytochrome-c oxidase (subunit III) <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''ctaE'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || cytochrome-c oxidase (subunit III) |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || respiration |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 23 kDa, 7.411 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 621 bp, 207 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ctaD]]'', ''[[ctaF]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB13364]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:ctaE_context.gif]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU14910 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 41: | Line 41: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ctaBCDEFG.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10217] |
=== Additional information=== | === Additional information=== | ||
| Line 52: | Line 52: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' 4 ferrocytochrome c + O<sub>2</sub> + 4 H<sup>+</sup> = 4 ferricytochrome c + 2 H<sub>2</sub>O (according to Swiss-Prot) |
| − | * '''Protein family:''' | + | * '''Protein family:''' cytochrome c oxidase subunit 3 family (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 70: | Line 70: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' |
| − | * '''Localization:''' | + | * '''Localization:''' cell membrane (according to Swiss-Prot) |
=== Database entries === | === Database entries === | ||
| Line 78: | Line 78: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P24012 P24012] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU14910] |
| − | * '''E.C. number:''' | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/1.9.3.1 1.9.3.1] |
=== Additional information=== | === Additional information=== | ||
| Line 88: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ | + | * '''Operon:''' ''[[ctaE]]'' |
| − | * '''[[Sigma factor]]:''' [[SigE]] | + | * '''[[Sigma factor]]:''' [[SigA]], [[SigE]] |
| − | * '''Regulation:''' | + | * '''Regulation:''' |
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| Line 113: | Line 113: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| − | |||
| − | |||
| − | |||
| − | |||
=Your additional remarks= | =Your additional remarks= | ||
| Line 122: | Line 118: | ||
=References= | =References= | ||
| − | |||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 17:32, 7 June 2009
- Description: cytochrome-c oxidase (subunit III)
| Gene name | ctaE |
| Synonyms | |
| Essential | no |
| Product | cytochrome-c oxidase (subunit III) |
| Function | respiration |
| MW, pI | 23 kDa, 7.411 |
| Gene length, protein length | 621 bp, 207 aa |
| Immediate neighbours | ctaD, ctaF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 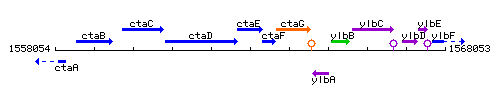
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU14910
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 4 ferrocytochrome c + O2 + 4 H+ = 4 ferricytochrome c + 2 H2O (according to Swiss-Prot)
- Protein family: cytochrome c oxidase subunit 3 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: P24012
- KEGG entry: [3]
- E.C. number: 1.9.3.1
Additional information
Expression and regulation
- Operon: ctaE
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed