Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' argininosuccinate lyase <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''argH'' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || argininosuccinate lyase |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of arginine | + | |style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of arginine |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 51 kDa, 4.859 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1383 bp, 461 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ytzD]]'', ''[[argG]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB14904]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | + | |colspan="2" | '''Genetic context''' <br/> [[Image:argH_context.gif]] | |
| − | |||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 31: | Line 29: | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/> | |
=The gene= | =The gene= | ||
| Line 37: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU29440 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 43: | Line 41: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' | + | * '''DBTBS entry:''' no entry |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG12571] |
=== Additional information=== | === Additional information=== | ||
| Line 54: | Line 52: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' N( | + | * '''Catalyzed reaction/ biological activity:''' 2-(N(omega)-L-arginino)succinate = fumarate + L-arginine (according to Swiss-Prot) |
| − | * '''Protein family:''' | + | * '''Protein family:''' Argininosuccinate lyase subfamily (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 80: | Line 78: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/O34858 O34858] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU29440] |
| − | * '''E.C. number:''' [http://www.expasy.org/enzyme/2. | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/4.3.2.1 4.3.2.1] |
=== Additional information=== | === Additional information=== | ||
| Line 90: | Line 88: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ | + | * '''Operon:''' ''[[argG]]-[[argH]]-[[ytzD]]'' |
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' repressed by casamino acids [http://www.ncbi.nlm.nih.gov/pubmed/12107147 PubMed], repressed by arginine ([[AhrC]]) | * '''Regulation:''' repressed by casamino acids [http://www.ncbi.nlm.nih.gov/pubmed/12107147 PubMed], repressed by arginine ([[AhrC]]) | ||
Revision as of 13:57, 8 June 2009
- Description: argininosuccinate lyase
| Gene name | argH |
| Synonyms | |
| Essential | no |
| Product | argininosuccinate lyase |
| Function | biosynthesis of arginine |
| MW, pI | 51 kDa, 4.859 |
| Gene length, protein length | 1383 bp, 461 aa |
| Immediate neighbours | ytzD, argG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 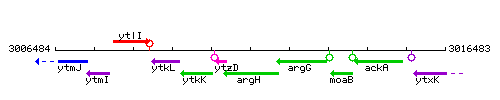
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU29440
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-(N(omega)-L-arginino)succinate = fumarate + L-arginine (according to Swiss-Prot)
- Protein family: Argininosuccinate lyase subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- Swiss prot entry: O34858
- KEGG entry: [2]
- E.C. number: 4.3.2.1
Additional information
Expression and regulation
- Regulatory mechanism: AhrC: transcription repression
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)