Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' replication initiation protein <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''dnaA'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''dnaH, dnaJ, dnaK '' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || yes [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || replication initiation protein |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || DNA replication |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 50 kDa, 6.035 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1338 bp, 446 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[rpmH]]'', ''[[dnaN]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB11777]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |colspan="2" | '''Genetic context''' <br/> [[Image:DnaA_dnaN_yaaA_recF_yaaB_gyrB_context.png]] |
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 35: | Line 35: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU00010 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | |||
| + | essential [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/dnaAN.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10065] |
=== Additional information=== | === Additional information=== | ||
| Line 54: | Line 56: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' dnaA family (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 70: | Line 72: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''Interactions:''' [[DnaA]]-[[YabA]], [[DnaA]]-[[YabA]]-[[DnaN]] [http://www.ncbi.nlm.nih.gov/sites/entrez/12060778 PubMed], [[DnaA]]-[[DnaD]] [http://www.ncbi.nlm.nih.gov/sites/entrez/11222620 PubMed] |
| − | * '''Localization:''' | + | * '''Localization:''' cytoplasm (according to Swiss-Prot), throughout the cytoplasm [http://www.ncbi.nlm.nih.gov/sites/entrez/10844689 PubMed] |
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' |
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P05648 P05648] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU00010] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
| Line 88: | Line 90: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ | + | * '''Operon:''' ''[[dnaA]]-[[dnaN]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/2987848 PubMed] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | '' | + | * '''[[Sigma factor]]:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/2987848 PubMed] |
| − | * '''Regulation:''' | + | * '''Regulation:''' negatively controlled by [[DnaA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/2168872 PubMed] and [[Spo0A]] [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] |
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' |
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 117: | Line 115: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | [[Philippe Noirot]], Jouy-en-Josas, France [http://locus.jouy.inra.fr/cms/index.php?id=18 homepage] | ||
| + | |||
| + | [[Peter Graumann]], Freiburg University, Germany [http://www.biologie.uni-freiburg.de/data/bio2/graumann/index.htm homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 122: | Line 123: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>10844689 11222620 12060778 16461910 2987848 2168872 14651647, </pubmed> |
| − | # | + | # Imai et al. (2000) Subcellular localization of Dna-initiation proteins of ''Bacillus subtilis'': evidence that chromosome replication begins at either edge of the nucleoids. ''Mol. Microbiol.'' '''36:''' 1037-1048. [http://www.ncbi.nlm.nih.gov/sites/entrez/10844689 PubMed] |
| − | # | + | # Ishigo-Oka et al. (2001) DnaD protein of ''Bacillus subtilis'' interacts with DnaA, the initiator protein of replication. ''J. Bacteriol.'' '''183:''' 2148-2150. [http://www.ncbi.nlm.nih.gov/sites/entrez/11222620 PubMed] |
| + | # Noirot-Gros et al. (2002) An expanded view of bacterial DNA replication. ''Proc. Natl. Acad. Sci. USA'' '''99:''' 8342-8347. [http://www.ncbi.nlm.nih.gov/sites/entrez/12060778 PubMed] | ||
| + | # Noirot-Gros et al. (2002) Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus Title ''Proc. Natl. Acad. Sci. USA'' '''103:''' 2368-2373. [http://www.ncbi.nlm.nih.gov/sites/entrez/16461910 PubMed] | ||
| + | # Ogasawara et al. (1985) Structure and function of the region of the replication origin of the ''Bacillus subtilis'' chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.''Nucl. Acids Res.'' '''11:''' 2267-2279. [http://www.ncbi.nlm.nih.gov/sites/entrez/2987848 PubMed] | ||
| + | # Fukuoka et al. (1990) Purification and characterization of an initiation protein for chromosomal replication, DnaA, in ''Bacillus subtilis''. ''J. Biochem.'' '''107:''' 732-739. [http://www.ncbi.nlm.nih.gov/sites/entrez/2168872 PubMed] | ||
| + | # Molle et al. (2003) The Spo0A regulon of ''Bacillus subtilis''.''Mol. Microbiol.'' '''50:''' 1683-1701. [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 14:01, 8 June 2009
- Description: replication initiation protein
| Gene name | dnaA |
| Synonyms | dnaH, dnaJ, dnaK |
| Essential | yes PubMed |
| Product | replication initiation protein |
| Function | DNA replication |
| MW, pI | 50 kDa, 6.035 |
| Gene length, protein length | 1338 bp, 446 aa |
| Immediate neighbours | rpmH, dnaN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 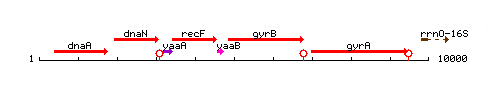
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU00010
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: dnaA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot), throughout the cytoplasm PubMed
Database entries
- Structure:
- Swiss prot entry: P05648
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Philippe Noirot, Jouy-en-Josas, France homepage
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Marie-Françoise Noirot-Gros, M Velten, M Yoshimura, S McGovern, T Morimoto, S D Ehrlich, N Ogasawara, P Polard, Philippe Noirot
Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2006, 103(7);2368-73
[PubMed:16461910]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Marie-Françoise Noirot-Gros, Etienne Dervyn, Ling Juan Wu, Peggy Mervelet, Jeffery Errington, S Dusko Ehrlich, Philippe Noirot
An expanded view of bacterial DNA replication.
Proc Natl Acad Sci U S A: 2002, 99(12);8342-7
[PubMed:12060778]
[WorldCat.org]
[DOI]
(P p)
D Ishigo-Oka, N Ogasawara, S Moriya
DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication.
J Bacteriol: 2001, 183(6);2148-50
[PubMed:11222620]
[WorldCat.org]
[DOI]
(P p)
Y Imai, N Ogasawara, D Ishigo-Oka, R Kadoya, T Daito, S Moriya
Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids.
Mol Microbiol: 2000, 36(5);1037-48
[PubMed:10844689]
[WorldCat.org]
[DOI]
(P p)
T Fukuoka, S Moriya, H Yoshikawa, N Ogasawara
Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis.
J Biochem: 1990, 107(5);732-9
[PubMed:2168872]
[WorldCat.org]
[DOI]
(P p)
N Ogasawara, S Moriya, H Yoshikawa
Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.
Nucleic Acids Res: 1985, 13(7);2267-79
[PubMed:2987848]
[WorldCat.org]
[DOI]
(P p)
- Imai et al. (2000) Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids. Mol. Microbiol. 36: 1037-1048. PubMed
- Ishigo-Oka et al. (2001) DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication. J. Bacteriol. 183: 2148-2150. PubMed
- Noirot-Gros et al. (2002) An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. USA 99: 8342-8347. PubMed
- Noirot-Gros et al. (2002) Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus Title Proc. Natl. Acad. Sci. USA 103: 2368-2373. PubMed
- Ogasawara et al. (1985) Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.Nucl. Acids Res. 11: 2267-2279. PubMed
- Fukuoka et al. (1990) Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 107: 732-739. PubMed
- Molle et al. (2003) The Spo0A regulon of Bacillus subtilis.Mol. Microbiol. 50: 1683-1701. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed