Difference between revisions of "Spo0A"
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || initiation of sporulation | |style="background:#ABCDEF;" align="center"|'''Function''' || initiation of sporulation | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/biofilm.html Biofilm], [http://subtiwiki.uni-goettingen.de/pathways/gene_regulation_nucleotides.html Nucleotides (regulation)], [http://subtiwiki.uni-goettingen.de/pathways/glutamate.html Ammonium/ glutamate], <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism],[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism], [http://subtiwiki.uni-goettingen.de/pathways/phosphorelay.html Phosphorelay], [http://subtiwiki.uni-goettingen.de/pathways/stress_response.html Stress], <br/>[http://subtiwiki.uni-goettingen.de/pathways/tRNA_charging.html tRNA charging],[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_synthesis.html Lipid synthesis] ''' | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/biofilm.html Biofilm], [http://subtiwiki.uni-goettingen.de/pathways/gene_regulation_nucleotides.html Nucleotides (regulation)], [http://subtiwiki.uni-goettingen.de/pathways/glutamate.html Ammonium/ glutamate], <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism],[http://subtiwiki.uni-goettingen.de/pathways/carbohydrate_metabolic_pathways.html Sugar catabolism], [http://subtiwiki.uni-goettingen.de/pathways/phosphorelay.html Phosphorelay], [http://subtiwiki.uni-goettingen.de/pathways/stress_response.html Stress], <br/>[http://subtiwiki.uni-goettingen.de/pathways/tRNA_charging.html tRNA charging],[http://subtiwiki.uni-goettingen.de/pathways/fatty_acid_synthesis.html Lipid synthesis], [http://subtiwiki.uni-goettingen.de/wiki/index.php/Protein_secretion Protein secretion] ''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 29 kDa, 5.989 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 29 kDa, 5.989 | ||
Revision as of 13:51, 11 December 2010
- Description: phosphorelay regulator, initiation of sporulation
| Gene name | spo0A |
| Synonyms | spo0C, spo0G, spoIIL, sof-1 |
| Essential | no |
| Product | phosphorelay response regulator |
| Function | initiation of sporulation |
| Metabolic function and regulation of this protein in SubtiPathways: Biofilm, Nucleotides (regulation), Ammonium/ glutamate, Central C-metabolism,Sugar catabolism, Phosphorelay, Stress, tRNA charging,Lipid synthesis, Protein secretion | |
| MW, pI | 29 kDa, 5.989 |
| Gene length, protein length | 801 bp, 267 aa |
| Immediate neighbours | yqiG, spoIVB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 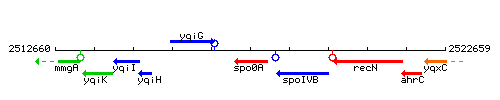
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription factors and their control, phosphorelay, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU24220
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Genes controlled by Spo0A
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: receives phosphorylation from Spo0B, dephosphorylation by Spo0E, direct phosphorylation by KinC upon potassium leakage PubMed
- Cofactor(s):
- Effectors of protein activity: the phosphorylation state affects DNA binding activity
- Localization:
Database entries
- Structure: 1QMP (with phosphorylated aspartate, Geobacillus stearothermophilus), 1FC3 (trans-activation domain, Geobacillus stearothermophilus), 1LQ1 (complex with DNA)
- UniProt: P06534
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: spo0A PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Tony Wilkinson, York University, U.K. homepage
- Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Reviews
José Eduardo González-Pastor
Cannibalism: a social behavior in sporulating Bacillus subtilis.
FEMS Microbiol Rev: 2011, 35(3);415-24
[PubMed:20955377]
[WorldCat.org]
[DOI]
(I p)
Michiel J L de Hoon, Patrick Eichenberger, Dennis Vitkup
Hierarchical evolution of the bacterial sporulation network.
Curr Biol: 2010, 20(17);R735-45
[PubMed:20833318]
[WorldCat.org]
[DOI]
(I p)
Tsutomu Katayama, Shogo Ozaki, Kenji Keyamura, Kazuyuki Fujimitsu
Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC.
Nat Rev Microbiol: 2010, 8(3);163-70
[PubMed:20157337]
[WorldCat.org]
[DOI]
(I p)
Daniel López, Roberto Kolter
Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis.
FEMS Microbiol Rev: 2010, 34(2);134-49
[PubMed:20030732]
[WorldCat.org]
[DOI]
(I p)
Daniel Schultz, Peter G Wolynes, Eshel Ben Jacob, José N Onuchic
Deciding fate in adverse times: sporulation and competence in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2009, 106(50);21027-34
[PubMed:19995980]
[WorldCat.org]
[DOI]
(I p)
S Seredick, G B Spiegelman
Lessons and questions from the structure of the Spo0A activation domain.
Trends Microbiol: 2001, 9(4);148-51
[PubMed:11286862]
[WorldCat.org]
[DOI]
(P p)
Original Publications