Difference between revisions of "HypO"
| Line 35: | Line 35: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[YybR (HypR) regulon]]}} | ||
| − | ''' | + | = The [[YybR (HypR) regulon]]:= |
| + | * ''[[yybR]]'' (hypR), ''[[yfkO]]'' (hypO) | ||
=The gene= | =The gene= | ||
| Line 45: | Line 47: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | hypO ohrA mutant more sensitive to NaOCl stress than ohrA single mutant | ||
=== Database entries === | === Database entries === | ||
Revision as of 11:04, 26 December 2011
- Description: NAD(P)H-flavin nitroreductase
| Gene name | yfkO (hypO) |
| Synonyms | hypO |
| Essential | no |
| Product | NAD(P)H-flavin nitroreductase |
| Function | probably involved in NaOCl and diamide detoxification |
| MW, pI | 25 kDa, 5.667 |
| Gene length, protein length | 663 bp, 221 aa |
| Immediate neighbours | treR, yfkN |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 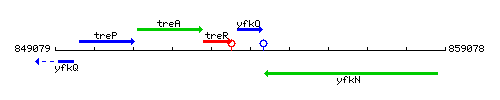
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The YybR (HypR) regulon:
The gene
Basic information
- Locus tag: BSU07830
Phenotypes of a mutant
hypO ohrA mutant more sensitive to NaOCl stress than ohrA single mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: nitroreductase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O34475
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: ykfO (hypO) (according to DBTBS)
- Regulation: induced under disulfide stress conditions (diamide, NaOCl)
- Regulatory mechanism: induction of YfkO (HypO) positively controlled by the MarR/DUF24-family regulator YybR (HypR)
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
G A Prosser, A V Patterson, D F Ackerley
uvrB gene deletion enhances SOS chromotest sensitivity for nitroreductases that preferentially generate the 4-hydroxylamine metabolite of the anti-cancer prodrug CB1954.
J Biotechnol: 2010, 150(1);190-4
[PubMed:20727918]
[WorldCat.org]
[DOI]
(I p)
Palm, Gottfried; Chi, Bui; Waack, Paul; Gronau, Katrin; Becher, Dörte; Albrecht, Dirk; Hinrichs, Winfried; Read, Randy; Antelmann, Haike
Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR
Nucleic Acid Research 2012, in press