Difference between revisions of "PnbA"
| Line 124: | Line 124: | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 745 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 745 {{PubMed|24696501}} | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 382 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 382 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 676 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 446 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 891 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
** GP951 (''pnbA''::''cat''), available in [[Stülke]] lab | ** GP951 (''pnbA''::''cat''), available in [[Stülke]] lab | ||
Revision as of 14:11, 17 April 2014
- Description: para-nitrobenzyl esterase
| Gene name | pnbA |
| Synonyms | estB |
| Essential | no |
| Product | para-nitrobenzyl esterase |
| Function | lipid degradation |
| Gene expression levels in SubtiExpress: pnbA | |
| MW, pI | 53 kDa, 4.776 |
| Gene length, protein length | 1467 bp, 489 aa |
| Immediate neighbours | slrR, padC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 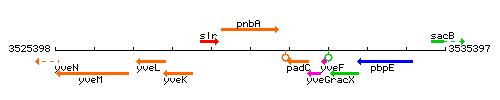
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
utilization of lipids, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34390
Phenotypes of a mutant
Database entries
- BsubCyc: BSU34390
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Triacylglycerol + H2O = diacylglycerol + a carboxylate (according to Swiss-Prot)
- Protein family: type-B carboxylesterase/lipase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-189 PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU34390
- UniProt: P37967
- KEGG entry: [2]
- E.C. number: 3.1.1.3 3.1.1.1]
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 745 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 382 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 676 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 446 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 891 PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References