Difference between revisions of "Rnz"
| Line 120: | Line 120: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 55 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 55 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 278 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 379 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 311 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
Revision as of 14:20, 17 April 2014
- Description: RNase Z
| Gene name | rnz |
| Synonyms | yqjK |
| Essential | yes PubMed |
| Product | endoribonuclease Z |
| Function | processing of CCA-less tRNA precursors |
| Gene expression levels in SubtiExpress: rnz | |
| MW, pI | 33 kDa, 5.805 |
| Gene length, protein length | 921 bp, 307 aa |
| Immediate neighbours | yqjL, zwf |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed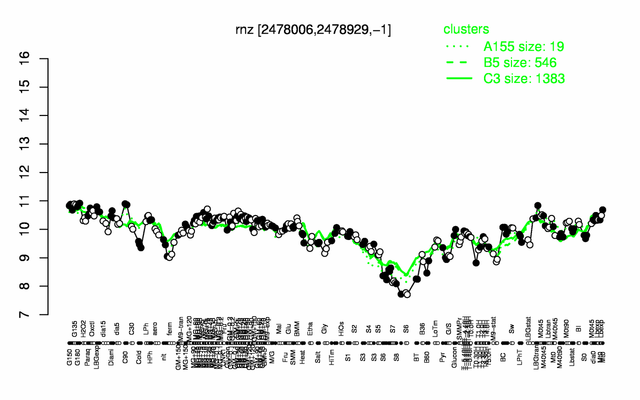
| |
Contents
Categories containing this gene/protein
Rnases, translation, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23840
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU23840
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Endonucleolytic cleavage of RNA, removing extra 3' nucleotides from tRNA precursor, generating 3' termini of tRNAs (according to Swiss-Prot)
- Protein family: RNase Z family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU23840
- UniProt: P54548
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: rnz (according to DBTBS)
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 55 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 278 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 379 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 311 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Ciaran Condon, IBPC, Paris, France Homepage
Your additional remarks
References
Reviews
Roland K Hartmann, Markus Gössringer, Bettina Späth, Susan Fischer, Anita Marchfelder
The making of tRNAs and more - RNase P and tRNase Z.
Prog Mol Biol Transl Sci: 2009, 85;319-68
[PubMed:19215776]
[WorldCat.org]
[DOI]
(P p)
B Späth, G Canino, A Marchfelder
tRNase Z: the end is not in sight.
Cell Mol Life Sci: 2007, 64(18);2404-12
[PubMed:17599240]
[WorldCat.org]
[DOI]
(P p)
Original Publications
Olivier Pellegrini, Inés Li de la Sierra-Gallay, Jérémie Piton, Laetitia Gilet, Ciarán Condon
Activation of tRNA maturation by downstream uracil residues in B. subtilis.
Structure: 2012, 20(10);1769-77
[PubMed:22940585]
[WorldCat.org]
[DOI]
(I p)
Alison Hunt, Joy P Rawlins, Helena B Thomaides, Jeff Errington
Functional analysis of 11 putative essential genes in Bacillus subtilis.
Microbiology (Reading): 2006, 152(Pt 10);2895-2907
[PubMed:17005971]
[WorldCat.org]
[DOI]
(P p)
Inés Li de la Sierra-Gallay, Nathalie Mathy, Olivier Pellegrini, Ciarán Condon
Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA.
Nat Struct Mol Biol: 2006, 13(4);376-7
[PubMed:16518398]
[WorldCat.org]
[DOI]
(P p)
Inés Li de la Sierra-Gallay, Olivier Pellegrini, Ciarán Condon
Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z.
Nature: 2005, 433(7026);657-61
[PubMed:15654328]
[WorldCat.org]
[DOI]
(I p)
Olivier Pellegrini, Jamel Nezzar, Anita Marchfelder, Harald Putzer, Ciarán Condon
Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis.
EMBO J: 2003, 22(17);4534-43
[PubMed:12941704]
[WorldCat.org]
[DOI]
(P p)