RpoE
- Description: RNA polymerase delta subunit, affects the regulation of RNA polymerase by the concentration of the initiating nucleoside triphosphate ([iNTP])
| Gene name | rpoE |
| Synonyms | |
| Essential | no |
| Product | RNA polymerase delta subunit |
| Function | transcription |
| Gene expression levels in SubtiExpress: rpoE | |
| Interactions involving this protein in SubtInteract: RpoE | |
| Metabolic function and regulation of this protein in SubtiPathways: rpoE | |
| MW, pI | 20 kDa, 3.654 |
| Gene length, protein length | 519 bp, 173 aa |
| Immediate neighbours | pyrG, acdA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed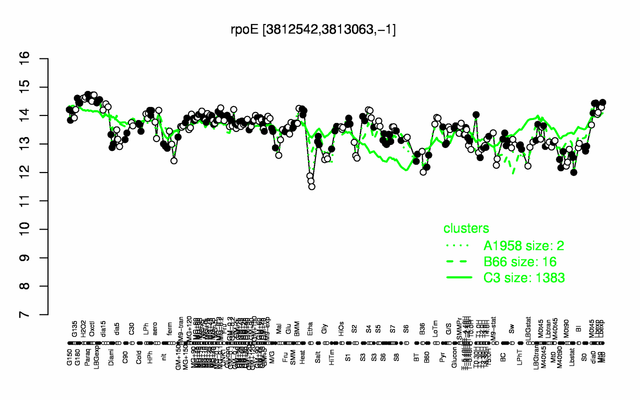
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU37160
Phenotypes of a mutant
Database entries
- BsubCyc: BSU37160
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: rpoE family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- closely associated to RNA polymerase (RpoA(2)-RpoB-RpoC) PubMed
- Localization:
- closely associated with RNA polymerase involved in transcribing both mRNA and rRNA operons PubMed
Database entries
- BsubCyc: BSU37160
- UniProt: P12464
- KEGG entry: [3]
- E.C. number: 2.7.7.6
Additional information
Expression and regulation
- Additional information:
- present at equimolar levels with RNA polymerase PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 965 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 3655 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Arthur Aronson, Purdue University, West Lafayette, USA homepage
Your additional remarks
References
Reviews
Lakshminarayan M Iyer, L Aravind
Insights from the architecture of the bacterial transcription apparatus.
J Struct Biol: 2012, 179(3);299-319
[PubMed:22210308]
[WorldCat.org]
[DOI]
(I p)
Original publications
Gabriel Demo, Veronika Papoušková, Jan Komárek, Pavel Kadeřávek, Olga Otrusinová, Pavel Srb, Alžbeta Rabatinová, Libor Krásný, Lukáš Žídek, Vladimír Sklenář, Michaela Wimmerová
X-ray vs. NMR structure of N-terminal domain of δ-subunit of RNA polymerase.
J Struct Biol: 2014, 187(2);174-186
[PubMed:24937760]
[WorldCat.org]
[DOI]
(I p)
Jana Wiedermannová, Petra Sudzinová, Tomaš Kovaľ, Alžbeta Rabatinová, Hana Šanderova, Olga Ramaniuk, Šimon Rittich, Jan Dohnálek, Zhihui Fu, Petr Halada, Peter Lewis, Libor Krásny
Characterization of HelD, an interacting partner of RNA polymerase from Bacillus subtilis.
Nucleic Acids Res: 2014, 42(8);5151-63
[PubMed:24520113]
[WorldCat.org]
[DOI]
(I p)
Veronika Papoušková, Pavel Kadeřávek, Olga Otrusinová, Alžbeta Rabatinová, Hana ŠSanderová, Jiří Nováček, Libor Krásný, Vladimír Sklenář, Lukáš Žídek
Structural study of the partially disordered full-length δ subunit of RNA polymerase from Bacillus subtilis.
Chembiochem: 2013, 14(14);1772-9
[PubMed:23868186]
[WorldCat.org]
[DOI]
(I p)
Alžbeta Rabatinová, Hana Šanderová, Jitka Jirát Matějčková, Jana Korelusová, Luděk Sojka, Ivan Barvík, Veronika Papoušková, Vladimír Sklenár, Lukáš Žídek, Libor Krásný
The δ subunit of RNA polymerase is required for rapid changes in gene expression and competitive fitness of the cell.
J Bacteriol: 2013, 195(11);2603-11
[PubMed:23543716]
[WorldCat.org]
[DOI]
(I p)
Anna Zawadzka-Kazimierczuk, Wiktor Koźmiński, Hana Sanderová, Libor Krásný
High dimensional and high resolution pulse sequences for backbone resonance assignment of intrinsically disordered proteins.
J Biomol NMR: 2012, 52(4);329-37
[PubMed:22350953]
[WorldCat.org]
[DOI]
(I p)
Jiří Nováček, Anna Zawadzka-Kazimierczuk, Veronika Papoušková, Lukáš Zídek, Hana Sanderová, Libor Krásný, Wiktor Koźmiński, Vladimír Sklenář
5D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion.
J Biomol NMR: 2011, 50(1);1-11
[PubMed:21424579]
[WorldCat.org]
[DOI]
(I p)
Veronika Motáčková, Jiří Nováček, Anna Zawadzka-Kazimierczuk, Krzysztof Kazimierczuk, Lukáš Zídek, Hana Sanderová, Libor Krásný, Wiktor Koźmiński, Vladimír Sklenář
Strategy for complete NMR assignment of disordered proteins with highly repetitive sequences based on resolution-enhanced 5D experiments.
J Biomol NMR: 2010, 48(3);169-77
[PubMed:20890634]
[WorldCat.org]
[DOI]
(I p)
Geoff P Doherty, Mark J Fogg, Anthony J Wilkinson, Peter J Lewis
Small subunits of RNA polymerase: localization, levels and implications for core enzyme composition.
Microbiology (Reading): 2010, 156(Pt 12);3532-3543
[PubMed:20724389]
[WorldCat.org]
[DOI]
(I p)
Veronika Motácková, Hana Sanderová, Lukás Zídek, Jirí Novácek, Petr Padrta, Alzbeta Svenková, Jana Korelusová, Jirí Jonák, Libor Krásný, Vladimír Sklenár
Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs.
Proteins: 2010, 78(7);1807-10
[PubMed:20310067]
[WorldCat.org]
[DOI]
(I p)
Hiroshi Matsuoka, Kazutake Hirooka, Yasutaro Fujita
Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation.
J Biol Chem: 2007, 282(8);5180-94
[PubMed:17189250]
[WorldCat.org]
[DOI]
(P p)
F J López de Saro, N Yoshikawa, J D Helmann
Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis.
J Biol Chem: 1999, 274(22);15953-8
[PubMed:10336502]
[WorldCat.org]
[DOI]
(P p)
F J López de Saro, A Y Woody, J D Helmann
Structural analysis of the Bacillus subtilis delta factor: a protein polyanion which displaces RNA from RNA polymerase.
J Mol Biol: 1995, 252(2);189-202
[PubMed:7545758]
[WorldCat.org]
[DOI]
(P p)
Y L Juang, J D Helmann
Pathway of promoter melting by Bacillus subtilis RNA polymerase at a stable RNA promoter: effects of temperature, delta protein, and sigma factor mutations.
Biochemistry: 1995, 34(26);8465-73
[PubMed:7599136]
[WorldCat.org]
[DOI]
(P p)
Y L Juang, J D Helmann
The delta subunit of Bacillus subtilis RNA polymerase. An allosteric effector of the initiation and core-recycling phases of transcription.
J Mol Biol: 1994, 239(1);1-14
[PubMed:7515111]
[WorldCat.org]
[DOI]
(P p)
M Lampe, C Binnie, R Schmidt, R Losick
Cloned gene encoding the delta subunit of Bacillus subtilis RNA polymerase.
Gene: 1988, 67(1);13-9
[PubMed:2843435]
[WorldCat.org]
[DOI]
(P p)
E I Hyde, M D Hilton, H R Whiteley
Interactions of Bacillus subtilis RNA polymerase with subunits determining the specificity of initiation. Sigma and delta peptides can bind simultaneously to core.
J Biol Chem: 1986, 261(35);16565-70
[PubMed:3097010]
[WorldCat.org]
(P p)
E C Achberger, H R Whiteley
The role of the delta peptide of the Bacillus subtilis RNA polymerase in promoter selection.
J Biol Chem: 1981, 256(14);7424-32
[PubMed:6788769]
[WorldCat.org]
(P p)