Difference between revisions of "CshA"
(→References) |
|||
| Line 171: | Line 171: | ||
=References= | =References= | ||
| − | + | <pubmed>16352840,20572937 , 12399512 16861794, 23175651,21803996 20709848,21710567</pubmed> | |
| − | <pubmed>16352840,20572937 , 12399512 16861794, 23175651,21803996</pubmed> | ||
== CshA in other organisms == | == CshA in other organisms == | ||
<pubmed> 23229022 21764917</pubmed> | <pubmed> 23229022 21764917</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:54, 17 November 2013
- Description: DEAD-box RNA helicase, important for adaptation to low temperatures
| Gene name | cshA |
| Synonyms | ydbR |
| Essential | no |
| Product | DEAD-box RNA helicase |
| Function | RNA helicase |
| Gene expression levels in SubtiExpress: cshA | |
| Interactions involving this protein in SubtInteract: CshA | |
| MW, pI | 57 kDa, 9.89 |
| Gene length, protein length | 1533 bp, 511 aa |
| Immediate neighbours | murF, ydbS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 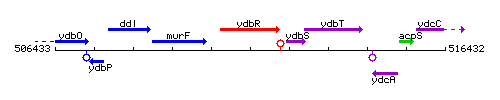
This image was kindly provided by SubtiList
| |
[None Expression at a glance] PubMed
| |
Contents
Categories containing this gene/protein
DEAD-box RNA helicases, translation, cold stress proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04580
Phenotypes of a mutant
- poor growth at low temperatures (16 to 20°C) PubMed
- reduced number of ribosomes PubMed
- no expression of the frlB-frlO-frlN-frlM-frlD operon PubMed
- strongly increased expression of the ysbA-ysbB operon PubMed
- transcription profile resulting from rny depletion: GEO PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: helicase C-terminal domain (according to Swiss-Prot) DEAD-box RNA helicase
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasma, colocalizes with the ribosomes PubMed, cell membrane PubMed
Database entries
- Structure:
- UniProt: P96614
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP1035 (aphA3), available in Jörg Stülke's lab
- GP1083 (cat), available in Jörg Stülke's lab
- Expression vector:
- for expression/ purification from B. subtilis with C-terminal Strep-tag, for SPINE, in pGP382: pGP1387, available in Jörg Stülke's lab
- for expression/ purification from B. subtilis with C-terminal Strep-tag, for SPINE, expression from the native chromomsomal site: GP1026 (aphA3), available in Jörg Stülke's lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP1386, available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- pGP1369 for chromosomal expression of CshA-YFP, available in Jörg Stülke's lab
- B. subtilis GP1081 cshA-gfp spc, available in Jörg Stülke's lab,
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- FLAG-tag construct:
- GP1010 (spc, based on pGP1331), available in Jörg Stülke's lab
- GP1074 (tet), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Mohamed Marahiel, Marburg University, Germany homepage
Your additional remarks
References
Martin Lehnik-Habrink, Leonie Rempeters, Ákos T Kovács, Christoph Wrede, Claudia Baierlein, Heike Krebber, Oscar P Kuipers, Jörg Stülke
DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other.
J Bacteriol: 2013, 195(3);534-44
[PubMed:23175651]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Joseph Newman, Fabian M Rothe, Alexandra S Solovyova, Cecilia Rodrigues, Christina Herzberg, Fabian M Commichau, Richard J Lewis, Jörg Stülke
RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli.
J Bacteriol: 2011, 193(19);5431-41
[PubMed:21803996]
[WorldCat.org]
[DOI]
(I p)
Olivier Delumeau, François Lecointe, Jan Muntel, Alain Guillot, Eric Guédon, Véronique Monnet, Michael Hecker, Dörte Becher, Patrice Polard, Philippe Noirot
The dynamic protein partnership of RNA polymerase in Bacillus subtilis.
Proteomics: 2011, 11(15);2992-3001
[PubMed:21710567]
[WorldCat.org]
[DOI]
(I p)
Franck Pandiani, Julien Brillard, Isabelle Bornard, Caroline Michaud, Stéphanie Chamot, Christophe Nguyen-the, Véronique Broussolle
Differential involvement of the five RNA helicases in adaptation of Bacillus cereus ATCC 14579 to low growth temperatures.
Appl Environ Microbiol: 2010, 76(19);6692-7
[PubMed:20709848]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Henrike Pförtner, Leonie Rempeters, Nico Pietack, Christina Herzberg, Jörg Stülke
The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex.
Mol Microbiol: 2010, 77(4);958-71
[PubMed:20572937]
[WorldCat.org]
[DOI]
(I p)
Yoshinari Ando, Kouji Nakamura
Bacillus subtilis DEAD protein YdbR possesses ATPase, RNA binding, and RNA unwinding activities.
Biosci Biotechnol Biochem: 2006, 70(7);1606-15
[PubMed:16861794]
[WorldCat.org]
[DOI]
(P p)
Karen Hunger, Carsten L Beckering, Frank Wiegeshoff, Peter L Graumann, Mohamed A Marahiel
Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis.
J Bacteriol: 2006, 188(1);240-8
[PubMed:16352840]
[WorldCat.org]
[DOI]
(P p)
Carsten L Beckering, Leif Steil, Michael H W Weber, Uwe Völker, Mohamed A Marahiel
Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis.
J Bacteriol: 2002, 184(22);6395-402
[PubMed:12399512]
[WorldCat.org]
[DOI]
(P p)
CshA in other organisms
Stella Oun, Peter Redder, Jean-Philippe Didier, Patrice François, Anna-Rita Corvaglia, Elena Buttazzoni, Caroline Giraud, Myriam Girard, Jacques Schrenzel, Patrick Linder
The CshA DEAD-box RNA helicase is important for quorum sensing control in Staphylococcus aureus.
RNA Biol: 2013, 10(1);157-65
[PubMed:23229022]
[WorldCat.org]
[DOI]
(I p)
Christelle M Roux, Jonathon P DeMuth, Paul M Dunman
Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex.
J Bacteriol: 2011, 193(19);5520-6
[PubMed:21764917]
[WorldCat.org]
[DOI]
(I p)