Difference between revisions of "DnaN"
| Line 55: | Line 55: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 89: | Line 86: | ||
** DnaN forms dimers {{PubMed|16045613}} | ** DnaN forms dimers {{PubMed|16045613}} | ||
| − | * '''Localization:''' | + | * '''Localization:''' |
| + | ** nucleoid (mid-cell spot) [http://www.ncbi.nlm.nih.gov/sites/entrez/9822387 PubMed] [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | ||
| + | ** forms large assemblies on DNA, called "clamp zones" {{PubMed|21419346}} | ||
=== Database entries === | === Database entries === | ||
| Line 140: | Line 139: | ||
<pubmed> 20157337 </pubmed> | <pubmed> 20157337 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| + | '''Additional publications:''' {{PubMed|21419346}} | ||
<pubmed>16942601 9822387 14651647,2987848 2168872 14651647 12060778 16461910, 16479537 19737352 19081080 11395445 20122408 20451384 15469515 16045613 </pubmed> | <pubmed>16942601 9822387 14651647,2987848 2168872 14651647 12060778 16461910, 16479537 19737352 19081080 11395445 20122408 20451384 15469515 16045613 </pubmed> | ||
'''Additional publications:''' {{PubMed|21050827}} | '''Additional publications:''' {{PubMed|21050827}} | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:04, 23 March 2011
- Description: DNA polymerase III (beta subunit), beta clamp, part of the replisome, also involved in DNA mismatch repair
| Gene name | dnaN |
| Synonyms | dnaG, dnaK |
| Essential | yes PubMed |
| Product | DNA polymerase III (beta subunit), beta clamp |
| Function | DNA replication, DNA repair |
| MW, pI | 41 kDa, 4.718 |
| Gene length, protein length | 1134 bp, 378 aa |
| Immediate neighbours | dnaA, yaaA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 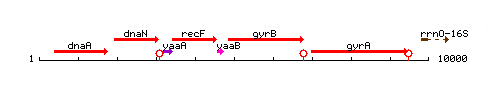
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
DNA replication, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00020
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1) (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P05649
- KEGG entry: [3]
- E.C. number: 2.7.7.7
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Philippe Noirot, Jouy-en-Josas, France homepage
Your additional remarks
References
Reviews
Tsutomu Katayama, Shogo Ozaki, Kenji Keyamura, Kazuyuki Fujimitsu
Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC.
Nat Rev Microbiol: 2010, 8(3);163-70
[PubMed:20157337]
[WorldCat.org]
[DOI]
(I p)
Original publications
Additional publications: PubMed
Peter T McKenney, Adam Driks, Haig A Eskandarian, Paul Grabowski, Jonathan Guberman, Katherine H Wang, Zemer Gitai, Patrick Eichenberger
A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat.
Curr Biol: 2010, 20(10);934-8
[PubMed:20451384]
[WorldCat.org]
[DOI]
(I p)
Glenn M Sanders, H Garry Dallmann, Charles S McHenry
Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases.
Mol Cell: 2010, 37(2);273-81
[PubMed:20122408]
[WorldCat.org]
[DOI]
(I p)
Alexi I Goranov, Adam M Breier, Houra Merrikh, Alan D Grossman
YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress.
Mol Microbiol: 2009, 74(2);454-66
[PubMed:19737352]
[WorldCat.org]
[DOI]
(I p)
Clarisse Defeu Soufo, Hervé Joël Defeu Soufo, Marie-Françoise Noirot-Gros, Astrid Steindorf, Philippe Noirot, Peter L Graumann
Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis.
Dev Cell: 2008, 15(6);935-41
[PubMed:19081080]
[WorldCat.org]
[DOI]
(I p)
Melanie B Berkmen, Alan D Grossman
Spatial and temporal organization of the Bacillus subtilis replication cycle.
Mol Microbiol: 2006, 62(1);57-71
[PubMed:16942601]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Marie-Françoise Noirot-Gros, M Velten, M Yoshimura, S McGovern, T Morimoto, S D Ehrlich, N Ogasawara, P Polard, Philippe Noirot
Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2006, 103(7);2368-73
[PubMed:16461910]
[WorldCat.org]
[DOI]
(P p)
Stéphane Duigou, S Dusko Ehrlich, Philippe Noirot, Marie-Françoise Noirot-Gros
DNA polymerase I acts in translesion synthesis mediated by the Y-polymerases in Bacillus subtilis.
Mol Microbiol: 2005, 57(3);678-90
[PubMed:16045613]
[WorldCat.org]
[DOI]
(P p)
Stéphane Duigou, S Dusko Ehrlich, Philippe Noirot, Marie-Françoise Noirot-Gros
Distinctive genetic features exhibited by the Y-family DNA polymerases in Bacillus subtilis.
Mol Microbiol: 2004, 54(2);439-51
[PubMed:15469515]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Marie-Françoise Noirot-Gros, Etienne Dervyn, Ling Juan Wu, Peggy Mervelet, Jeffery Errington, S Dusko Ehrlich, Philippe Noirot
An expanded view of bacterial DNA replication.
Proc Natl Acad Sci U S A: 2002, 99(12);8342-7
[PubMed:12060778]
[WorldCat.org]
[DOI]
(P p)
Y Ogura, Y Imai, N Ogasawara, S Moriya
Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis.
J Bacteriol: 2001, 183(13);3833-41
[PubMed:11395445]
[WorldCat.org]
[DOI]
(P p)
K P Lemon, A D Grossman
Localization of bacterial DNA polymerase: evidence for a factory model of replication.
Science: 1998, 282(5393);1516-9
[PubMed:9822387]
[WorldCat.org]
[DOI]
(P p)
T Fukuoka, S Moriya, H Yoshikawa, N Ogasawara
Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis.
J Biochem: 1990, 107(5);732-9
[PubMed:2168872]
[WorldCat.org]
[DOI]
(P p)
N Ogasawara, S Moriya, H Yoshikawa
Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames.
Nucleic Acids Res: 1985, 13(7);2267-79
[PubMed:2987848]
[WorldCat.org]
[DOI]
(P p)
Additional publications: PubMed