Difference between revisions of "FbaA"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/><br/><br/><br/><br/> | <br/><br/><br/><br/><br/><br/> | ||
| Line 123: | Line 119: | ||
* '''Sigma factor:''' | * '''Sigma factor:''' | ||
| − | * '''Regulation:''' constitutively expressed {{PubMed|11489127}} | + | * '''Regulation:''' |
| + | ** constitutively expressed {{PubMed|11489127}} | ||
| + | ** strongly repressed in response to glucose starvation in M9 medium {{PubMed|23033921}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| Line 154: | Line 152: | ||
=References= | =References= | ||
| − | + | '''Additional publications:''' {{PubMed|23033921}} | |
<pubmed>17218307, 15125960, 24624 16843441 11489127 20525796</pubmed> | <pubmed>17218307, 15125960, 24624 16843441 11489127 20525796</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:34, 8 October 2012
- Description: fructose 1,6-bisphosphate aldolase, glycolytic/ gluconeogenic enzyme
| Gene name | fbaA |
| Synonyms | fba, fba1, tsr |
| Essential | yes |
| Product | fructose-1,6-bisphosphate aldolase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| Gene expression levels in SubtiExpress: fbaA | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism, Sugar catabolism | |
| MW, pI | 30,2 kDa, 5.03 |
| Gene length, protein length | 855 bp, 285 amino acids |
| Immediate neighbours | ywjH, spo0F |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 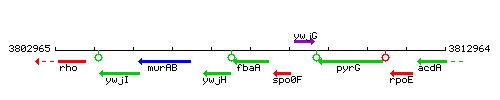
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU37120
Phenotypes of a mutant
- Essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-fructose 1,6-bisphosphate = glycerone phosphate + D-glyceraldehyde 3-phosphate (according to Swiss-Prot)
- Protein family: class II fructose-bisphosphate aldolase family (according to Swiss-Prot)
- Paralogous protein(s): FbaB
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten PubMed
- Domains:
- 2 x Dihydroxyacetone phosphate binding domain (210–212), (231–234)
- Modification: phosphorylation on Thr-212 and Thr-234 PubMed
- Cofactor(s): Zn2+ (Metalloenzyme)
- Effectors of protein activity:
Database entries
- Structure: 3Q94 (from Bacillus anthracis)
- UniProt: P13243
- KEGG entry: [3]
- E.C. number: 4.1.2.13
Additional information
- Binds 2 zinc ions per subunit. One is catalytic and the other provides a structural contribution
- extensive information on the structure and enzymatic properties of FbaA can be found at Proteopedia
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
- The mRNA has a long 5' leader region. This may indicate RNA-based regulation PubMed
Biological materials
- Expression vector:
- for expression in B. subtilis, in pBQ200: pGP1423, available in Stülke lab
- for expression/ purification from B. subtilis with N-terminal Strep-tag, for SPINE, in pGP380: pGP88, available in Stülke lab
- for expression/ purification from E. coli with N-terminal His-tag, in pWH844: pGP395, available in Stülke lab
- lacZ fusion: pGP601 (in pAC6)
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Jun Hyuck Lee, Jungdon Bae, Dooil Kim, Yongseok Choi, Young Jun Im, Sukhoon Koh, Joong Su Kim, Mun-Kyoung Kim, Gil Bu Kang, Suk-In Hong, Dae-Sil Lee, Soo Hyun Eom
Stereoselectivity of fructose-1,6-bisphosphate aldolase in Thermus caldophilus.
Biochem Biophys Res Commun: 2006, 347(3);616-25
[PubMed:16843441]
[WorldCat.org]
[DOI]
(P p)
Matthieu Fonvielle, Philippe Weber, Kasia Dabkowska, Michel Therisod
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases.
Bioorg Med Chem Lett: 2004, 14(11);2923-6
[PubMed:15125960]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
S Ujita
Fructose 1,6-bisphosphate aldolases from spores and vegetative cells of Bacillus subtilis PCI 219.
J Biochem: 1978, 83(2);493-502
[PubMed:24624]
[WorldCat.org]
[DOI]
(P p)