Difference between revisions of "PcrB"
Raphael2215 (talk | contribs) |
|||
| Line 70: | Line 70: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** production of heptaprenylglyceryl phosphate from heptaprenyl diphosphate and glycerol-1-phosphate {{PubMed|23322418}} | ||
* '''Protein family:''' GGGP synthase family (according to Swiss-Prot) | * '''Protein family:''' GGGP synthase family (according to Swiss-Prot) | ||
| Line 94: | Line 95: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1VIZ 1VIZ] | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1VIZ 1VIZ] {{PubMed|23322418}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O34790 O34790] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O34790 O34790] | ||
| Line 137: | Line 138: | ||
=References= | =References= | ||
| − | <pubmed>9701819 22614947 21761520 18558723 </pubmed> | + | <pubmed>9701819 22614947 21761520 18558723 23322418 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:08, 17 January 2013
- Description: heptaprenylglyceryl phosphate synthase
| Gene name | pcrB |
| Synonyms | yerE |
| Essential | no |
| Product | heptaprenylglyceryl phosphate synthase |
| Function | unknown |
| Gene expression levels in SubtiExpress: pcrB | |
| MW, pI | 24 kDa, 4.204 |
| Gene length, protein length | 684 bp, 228 aa |
| Immediate neighbours | yerD, pcrA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 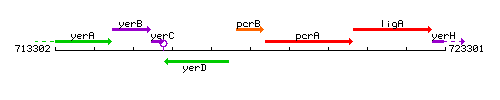
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06600
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- production of heptaprenylglyceryl phosphate from heptaprenyl diphosphate and glycerol-1-phosphate PubMed
- Protein family: GGGP synthase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- forms dimers PubMed
Database entries
- UniProt: O34790
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Feifei Ren, Xinxin Feng, Tzu-Ping Ko, Chun-Hsiang Huang, Yumei Hu, Hsiu-Chien Chan, Yi-Liang Liu, Ke Wang, Chun-Chi Chen, Xuefei Pang, Miao He, Yujie Li, Eric Oldfield, Rey-Ting Guo
Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus.
Chembiochem: 2013, 14(2);195-9
[PubMed:23322418]
[WorldCat.org]
[DOI]
(I p)
David Peterhoff, Hermann Zellner, Harald Guldan, Rainer Merkl, Reinhard Sterner, Patrick Babinger
Dimerization determines substrate specificity of a bacterial prenyltransferase.
Chembiochem: 2012, 13(9);1297-303
[PubMed:22614947]
[WorldCat.org]
[DOI]
(I p)
Harald Guldan, Frank-Michael Matysik, Marco Bocola, Reinhard Sterner, Patrick Babinger
Functional assignment of an enzyme that catalyzes the synthesis of an archaea-type ether lipid in bacteria.
Angew Chem Int Ed Engl: 2011, 50(35);8188-91
[PubMed:21761520]
[WorldCat.org]
[DOI]
(I p)
Harald Guldan, Reinhard Sterner, Patrick Babinger
Identification and characterization of a bacterial glycerol-1-phosphate dehydrogenase: Ni(2+)-dependent AraM from Bacillus subtilis.
Biochemistry: 2008, 47(28);7376-84
[PubMed:18558723]
[WorldCat.org]
[DOI]
(I p)
M A Petit, E Dervyn, M Rose, K D Entian, S McGovern, S D Ehrlich, C Bruand
PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication.
Mol Microbiol: 1998, 29(1);261-73
[PubMed:9701819]
[WorldCat.org]
[DOI]
(P p)