Difference between revisions of "Tpi"
| Line 44: | Line 44: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[carbon core metabolism]]}}, | {{SubtiWiki category|[[carbon core metabolism]]}}, | ||
| − | {{SubtiWiki category|[[phosphoproteins]]}} | + | {{SubtiWiki category|[[phosphoproteins]]}}, |
| + | [[most abundant proteins]] | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 81: | Line 82: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylation on Ser-213 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | * '''Modification:''' phosphorylation on Ser-213 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' inhibited by 2-phosphoglycolate (in ''B. stearothermophilus'') [http://www.ncbi.nlm.nih.gov/sites/entrez/8580851 PubMed] | * '''Effectors of protein activity:''' inhibited by 2-phosphoglycolate (in ''B. stearothermophilus'') [http://www.ncbi.nlm.nih.gov/sites/entrez/8580851 PubMed] | ||
| Line 126: | Line 127: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
=Biological materials = | =Biological materials = | ||
| Line 150: | Line 152: | ||
=References= | =References= | ||
| − | <pubmed>12850135 11489127 17505547 8021172 17218307 8580851 19193632 21821766 23420519</pubmed> | + | <pubmed>12850135 11489127 17505547 8021172 17218307 8580851 19193632 21821766 23420519 15378759</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:37, 5 March 2014
- Description: triose phosphate isomerase, glycolytic/ gluconeogenic enzyme
| Gene name | tpi |
| Synonyms | tpiA |
| Essential | no |
| Product | triosephosphate isomerase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| Gene expression levels in SubtiExpress: tpi | |
| Interactions involving this protein in SubtInteract: Tpi | |
| Metabolic function and regulation of this protein in SubtiPathways: tpi | |
| MW, pI | 26,9 kDa, 4.79 |
| Gene length, protein length | 759 bp, 253 amino acids |
| Immediate neighbours | pgm, pgk |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 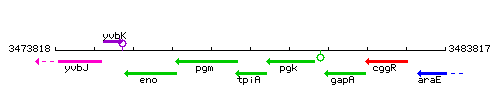
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed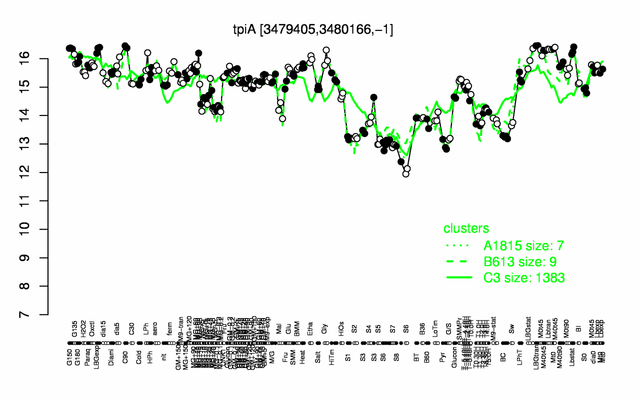
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33920
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: D-glyceraldehyde 3-phosphate = dihydroxyacetone phosphate (according to Swiss-Prot)
- Protein family: triosephosphate isomerase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylation on Ser-213 PubMed
- Effectors of protein activity: inhibited by 2-phosphoglycolate (in B. stearothermophilus) PubMed
- Localization:
- cytoplasm PubMed
Database entries
- Structure: 1BTM (complex with 2-phosphoglycolic acid, Geobacillus stearothermophilus), complex with 2-phosphpoglycolic acid, Geobacillus stearothermophilus NCBI
- UniProt: P27876
- KEGG entry: [3]
- E.C. number: 5.3.1.1
Additional information
- extensive information on the structure and enzymatic properties of Tpi can be found at Proteopedia
Expression and regulation
- Regulation:
- Additional information:
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant: GP700 (cat), available in Jörg Stülke's lab, PubMed
- Expression vector:
- pGP394 (N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- pGP89 (N-terminal Strep-tag, for SPINE, expression in B. subtilis), available in Jörg Stülke's lab
- pGP1511 (expression in B. subtilis, in pBQ200), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Fabian M Commichau, Nico Pietack, Jörg Stülke
Essential genes in Bacillus subtilis: a re-evaluation after ten years.
Mol Biosyst: 2013, 9(6);1068-75
[PubMed:23420519]
[WorldCat.org]
[DOI]
(I p)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Laurent Jannière, Danielle Canceill, Catherine Suski, Sophie Kanga, Bérengère Dalmais, Roxane Lestini, Anne-Françoise Monnier, Jérôme Chapuis, Alexander Bolotin, Marina Titok, Emmanuelle Le Chatelier, S Dusko Ehrlich
Genetic evidence for a link between glycolysis and DNA replication.
PLoS One: 2007, 2(5);e447
[PubMed:17505547]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
L F Delboni, S C Mande, F Rentier-Delrue, V Mainfroid, S Turley, F M Vellieux, J A Martial, W G Hol
Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions.
Protein Sci: 1995, 4(12);2594-604
[PubMed:8580851]
[WorldCat.org]
[DOI]
(P p)
M A Leyva-Vazquez, P Setlow
Cloning and nucleotide sequences of the genes encoding triose phosphate isomerase, phosphoglycerate mutase, and enolase from Bacillus subtilis.
J Bacteriol: 1994, 176(13);3903-10
[PubMed:8021172]
[WorldCat.org]
[DOI]
(P p)