Difference between revisions of "CcpA"
(→Categories containing this gene/protein) |
|||
| Line 45: | Line 45: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[transcription factors and their control]]}}, | + | * {{SubtiWiki category|[[transcription factors and their control]]}}, |
{{SubtiWiki category|[[regulators of core metabolism]]}} | {{SubtiWiki category|[[regulators of core metabolism]]}} | ||
| + | * see also: [[glutamate metabolism]] | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
Revision as of 18:14, 3 June 2012
- Description: Carbon catabolite control protein A, involved in glucose regulation of many genes; represses catabolic genes and activates genes involved in excretion of excess carbon
| Gene name | ccpA |
| Synonyms | graR, alsA, amyR |
| Essential | no |
| Product | transcriptional regulator (LacI family) |
| Function | mediates carbon catabolite repression (CCR) |
| Interactions involving this protein in SubtInteract: CcpA | |
| Metabolic function and regulation of this protein in SubtiPathways: Nucleoside catabolism, Nucleotides (regulation), Ile, Leu, Val, His, Coenzyme A, Central C-metabolism | |
| MW, pI | 36,8 kDa, 5.06 |
| Gene length, protein length | 1002 bp, 334 amino acids |
| Immediate neighbours | motP, aroA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 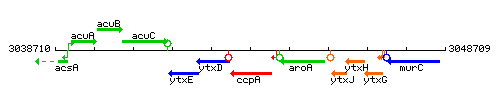
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
- 1 Categories containing this gene/protein
- 2 This gene is a member of the following regulons
- 3 The CcpA regulon
- 4 The gene
- 5 The protein
- 6 Expression and regulation
- 7 Biological materials
- 8 Labs working on this gene/protein
- 9 Your additional remarks
- 10 References
- 10.1 Reviews
- 10.2 General and physiological studies
- 10.3 Global analyses (proteome, transcriptome, ChIP-chip)
- 10.4 Repression of target genes by CcpA
- 10.5 Positive regulation of gene expression by CcpA
- 10.6 Control of CcpA activity
- 10.7 CcpA-DNA interaction
- 10.8 Functional analysis of CcpA
- 10.9 Structural analyses
Categories containing this gene/protein
- see also: glutamate metabolism
This gene is a member of the following regulons
The CcpA regulon
The gene
Basic information
- Locus tag: BSU29740
Phenotypes of a mutant
Loss of carbon catabolite repression. Loss of PTS-dependent sugar transport due to excessive phosphorylation of HPr by HprK. The mutant is unable to grow on a minimal medium with glucose and ammonium as the only sources of carbon and nitrogen, respectively.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcriptional regulator of carbon catabolite repression (CCR)
- Protein family: LacI family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- HTH LacI-type Domain (1 – 58)
- DNA binding Domain (6 – 25)
- Modification:
- Effectors of protein activity:glucose-6-phosphate, fructose-1,6-bisphosphate Pubmed
Database entries
- Structure:
- 2JCG (Apoprotein from Bacillus megaterium)
- CcpA-Crh-DNA-complex NCBI
- complex with P-Ser-HPr and sulphate ions NCBI
- 3OQM (complex of B. subtilis CcpA with P-Ser-HPr and the ackA operator site)
- 3OQN (complex of B. subtilis CcpA with P-Ser-HPr and the gntR operator site)
- 3OQO (complex of B. subtilis CcpA with P-Ser-HPr and a optimal synthetic operator site)
- UniProt: P25144
- KEGG entry: [3]
Additional information
Expression and regulation
- Sigma factor:
- Regulation: constitutively expressed PubMed
- Additional information: there are about 3.000 molecules of CcpA per cell PubMed, this corresponds to a concentration of 3 myM (according to PubMed)
Biological materials
- Mutant: QB5407 (spc), GP302 (erm), GP300 (an in frame deletion of ccpA), available in Stülke lab; WH649 (aphA3), available in Gerald Seidel's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
Labs working on this gene/protein
- Gerald Seidel, Erlangen University, Germany Homepage
- Richard Brennan, Houston, Texas, USA Homepage
- Milton H. Saier, University of California at San Diego, USA Homepage
- Yasutaro Fujita, University of Fukuyama, Japan
- Jörg Stülke, University of Göttingen, Germany Homepage
- Oscar Kuipers, University of Groningen, The Netherlands Homepage
Your additional remarks
References
Reviews
General and physiological studies
Additional publications: PubMed
Global analyses (proteome, transcriptome, ChIP-chip)
Repression of target genes by CcpA
Additional publications: PubMed
José Manuel Inácio, Isabel de Sá-Nogueira
trans-Acting factors and cis elements involved in glucose repression of arabinan degradation in Bacillus subtilis.
J Bacteriol: 2007, 189(22);8371-6
[PubMed:17827291]
[WorldCat.org]
[DOI]
(I p)
Soo-Keun Choi, Milton H Saier
Mechanism of CcpA-mediated glucose repression of the resABCDE operon of Bacillus subtilis.
J Mol Microbiol Biotechnol: 2006, 11(1-2);104-10
[PubMed:16825793]
[WorldCat.org]
[DOI]
(P p)
Soo-Keun Choi, Milton H Saier
Regulation of pho regulon gene expression by the carbon control protein A, CcpA, in Bacillus subtilis.
J Mol Microbiol Biotechnol: 2005, 10(1);40-50
[PubMed:16491025]
[WorldCat.org]
[DOI]
(P p)
Soo-Keun Choi, Milton H Saier
Regulation of sigL expression by the catabolite control protein CcpA involves a roadblock mechanism in Bacillus subtilis: potential connection between carbon and nitrogen metabolism.
J Bacteriol: 2005, 187(19);6856-61
[PubMed:16166551]
[WorldCat.org]
[DOI]
(P p)
Boris R Belitsky, Hyun-Jin Kim, Abraham L Sonenshein
CcpA-dependent regulation of Bacillus subtilis glutamate dehydrogenase gene expression.
J Bacteriol: 2004, 186(11);3392-8
[PubMed:15150224]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Agnes Roux, Abraham L Sonenshein
Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes.
Mol Microbiol: 2002, 45(1);179-90
[PubMed:12100558]
[WorldCat.org]
[DOI]
(P p)
Hyun-Jin Kim, Cécile Jourlin-Castelli, Sam-In Kim, Abraham L Sonenshein
Regulation of the bacillus subtilis ccpC gene by ccpA and ccpC.
Mol Microbiol: 2002, 43(2);399-410
[PubMed:11985717]
[WorldCat.org]
[DOI]
(P p)
Emmanuelle Darbon, Pascale Servant, Sandrine Poncet, Josef Deutscher
Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpFK expression.
Mol Microbiol: 2002, 43(4);1039-52
[PubMed:11929549]
[WorldCat.org]
[DOI]
(P p)
I Martin-Verstraete, J Stülke, A Klier, G Rapoport
Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon.
J Bacteriol: 1995, 177(23);6919-27
[PubMed:7592486]
[WorldCat.org]
[DOI]
(P p)
F J Grundy, A J Turinsky, T M Henkin
Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA.
J Bacteriol: 1994, 176(15);4527-33
[PubMed:7913927]
[WorldCat.org]
[DOI]
(P p)
Positive regulation of gene expression by CcpA
Control of CcpA activity
CcpA-DNA interaction
Functional analysis of CcpA
Structural analyses
Bernhard Loll, Wolfram Saenger, Jacek Biesiadka
Structure of full-length transcription regulator CcpA in the apo form.
Biochim Biophys Acta: 2007, 1774(6);732-6
[PubMed:17500051]
[WorldCat.org]
[DOI]
(P p)
Rajesh Kumar Singh, Gottfried J Palm, Santosh Panjikar, Winfried Hinrichs
Structure of the apo form of the catabolite control protein A (CcpA) from Bacillus megaterium with a DNA-binding domain.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2007, 63(Pt 4);253-7
[PubMed:17401189]
[WorldCat.org]
[DOI]
(I p)
Maria A Schumacher, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate.
J Mol Biol: 2007, 368(4);1042-50
[PubMed:17376479]
[WorldCat.org]
[DOI]
(P p)
Vincent Chaptal, Virginie Gueguen-Chaignon, Sandrine Poncet, Cécile Lecampion, Philippe Meyer, Josef Deutscher, Anne Galinier, Sylvie Nessler, Solange Moréra
Structural analysis of B. subtilis CcpA effector binding site.
Proteins: 2006, 64(3);814-6
[PubMed:16755587]
[WorldCat.org]
[DOI]
(I p)
Maria A Schumacher, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J Biol Chem: 2006, 281(10);6793-800
[PubMed:16316990]
[WorldCat.org]
[DOI]
(P p)
Maria A Schumacher, Gregory S Allen, Marco Diel, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell: 2004, 118(6);731-41
[PubMed:15369672]
[WorldCat.org]
[DOI]
(P p)
J Tebbe, P Orth, E K Küster-Schöck, W Hillen, W Saenger, W Hinrichs
Crystallization and preliminary X-ray analyses of catabolite control protein A, free and in complex with its DNA-binding site.
Acta Crystallogr D Biol Crystallogr: 2000, 56(Pt 1);67-9
[PubMed:10666630]
[WorldCat.org]
[DOI]
(P p)